DataseamGrid Finds “Holy Grail” of Cancer Treatment
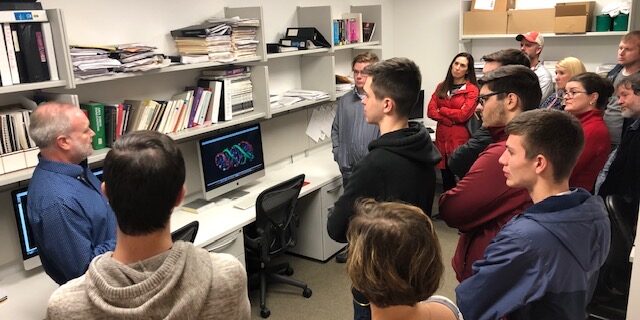
Dr. John Trent talks to school technicians about results on DataseamGrid. (Louisville, Ky) – Nearly ten years ago John Trent Ph.D. of the Molecular Modeling Facility at UofL used the combined power of thousands of computers in classrooms across the state of Kentucky to discover potential compounds that stop cancer from growing. The University recently announced…